Open Journal of
Clinical and Medical Images
Research Article - Open Access, Volume 1
Screening of hub-genes, key pathways, and targeted medications in laryngeal squamous cell carcinoma using bioinformatics analysis
Yuan Liang; Lei Zhao*
Department of Otorhinolaryngology, Affiliated Hospital of Chengde Medical College, Chengde, Hebei province, 067000, China.
*Corresponding Author: Lei Zhao
Department of Otorhinolaryngology, Affiliated Hospital
of Hebei University, No. 212, Yuhua East Road, Baoding,
071000, China.
Email: erbiyanhouxue@126.com
Received : Oct 03, 2021
Accepted : Nov 08, 2021
Published : Nov 12, 2021
Archived : www.jclinmedimages.org
Copyright : © Zhao L (2021).
Abstract
For Identifying the hub-genes and targeted medications for regulating laryngeal squamous cell carcinoma (LSCC). Differently expressed mRNAs (DEMs) in LSCC were acquired from Gene Expression Omnibus (GEO) by GEO2R. Also, the hub-genes of LSCC were calculated with cyto Hubba and validated by The Cancer Genome Atlas (TCGA). Then, functional enrichment analyses, diagnostic and prognostic values, and therapeutic medications of hub-genes were explored. Subsequently, GSE51985, GSE84957, and GSE143224 were screened out from GEO. Accordingly, 515, 1720, and 848 DEMs were filtered from them, respectively. Total 34 up-regulated and 42 down-regulated DEMs, the intersections of the three datasets, were used for identifying the hub-genes. Finally, 10 up- and 10 down-regulated DEMs were defined as hub-genes and verified by TCGA. Followed analyses indicated that the hub-genes participated in the regulations of PI3K-Akt signaling pathway, metabolic pathways, pathways in cancer, and other signaling pathways and molecular functions. Moreover, certain hub-genes (LAMC2 and SPP1) presented potential diagnostic and prognostic values. Even, certain drugs may interact with these hub-genes. Meaningfully, the present study indicated that the hub-genes of LSCC involved in the pathogenesis of LSCC through PI3K-Akt signaling pathway, metabolic pathways, and pathways in cancer. Even, LAMC2 and SPP1 may be employed as the special markers for LSCC diagnosis or prognosis and may act as therapeutic targets for certain drugs (ASK-8007, Ocriplasmin or Calcitonin).
Keywords: laryngeal tumor; hub-gene; target therapy; TCGA; GEO; expression profile.
Citation: Liang Y, Zhao L. Screening of hub-genes, key pathways, and targeted medications in laryngeal squamous cell carcinoma using bioinformatics analysis. Open J Clin Med Images. 2021; 1(1): 1006
Introduction
Laryngeal cancer is a multiple histological types of malignant tumor and around 98% of the malignancies are laryngeal squamous cell carcinomas (LSCC) [1]. The morbidity of laryngeal cancer is around at 2.76 cases/100,000 populations/year and has increased by 12% for the last three decades [2]. Meanwhile, the mortality of laryngeal cancer is around at 1.66 deaths/100,000 populations/year and has decreased by 5% [2]. Approximately 97% of the laryngeal cancer occurred in males and roughly 71% aged 51~70 years [1]. The five-year relative survival rate of laryngeal cancer is around 63% [3], but over 80% of the laryngeal cancer patients have the chance to survive for more than five years after undergoing radical surgery and systematic treatments [1]. However, the ascending in morbidity and descending in mortality means an increasing prevalence. In 2020, 177,422 new cases of laryngeal cancer are reckoned to occur in worldwide [4]. Furthermore, around 98% of the laryngeal cancer patients are diagnosed in advanced clinical stages (III~IV) [1]. All of these indicate that most of these patients will endure the laryngeal functional changes, even loss.
The heavy burden of laryngeal cancer directs researchers to explore the pathogenesis of LSCC, and to screen specific tumor markers and effective molecular targets. Prior studies have verified that multiple molecules participated in the occurrence and development of LSCC through diverse biological functions and mechanisms, involving genomics [5], transcriptomics [6], proteomics [7], and epigenetics [8]. For instance, circCORO1C, an over-expressed circular RNA in LSCC, accelerates the malignancy progression and the poor prognosis of LSCC patients through regulating the let-7c-5p/PBX3 pathway [9]. And, long non-coding RNA RP11-159K7.2 advances the proliferation and invasion ability of LSCC through up-regulating DNMT3A expression by complementary binding to miR-206, and shows the prognostic value for LSCC patients [10]. Additionally, certain functional proteins, such as FADS1, FOXJ1, and MMP2/3 also participate in the biological processes of LSCC by governing AKT/mTOR, Wnt/βcatenin, and PI3K/AKT-NF-κB pathways, respectively [11-13]. All of above studies suggest that it is feasible and significant to explore the potential biomarkers of LSCC.
Hence, the present study aimed to screen out the critical regulatory molecules that involved in the initiation and progression of LSCC through integrated analysis of multiple expression profiles. Consequently, the study identified and validated 10 upand 10 down-regulated hub-genes that affected the evolution of LSCC through PI3K-Akt and metabolic pathways, even other pivotal biological processes. Further analyses indicated the potential diagnostic and prognostic values of LAMC2 and SPP1 for LSCC. In addition, the study also preliminarily explored the therapeutic drugs interact with LAMC2 and SPP1. Significantly, the finding of the present study projected valuable references for the screening of specific tumor markers and effective targets for the diagnosis, prognosis, and target therapy of LSCC. The detailed analyses are as follows.
Materials & Methods
Retrieving and filtering of expression profiles: According to the retrieval filters (Study type: Expression profiling by array and Attribute name: tissue) and search terms: LSCC OR “laryn geal squamous cell carcinoma”, LSCC related mRNA expression profiles were retrieved from Gene Expression Ominibus (GEO) DataSets (http://www.ncbi.nlm.nih.gov/geo/), a free repository tool for querying and downloading expression profiles [14]. The retrieved profiles were further filtered according to the screen ing criteria: (1) mRNA profiles; (2) data deriving from cancerous tissues and paired normal tissues; (3) retrieval period: from ini tial to May 04, 2020, the retrieval date.
Screening of differential expressed genes: Based on the de¬fault false discovery rate (Benjamini & Hochberg) and P<0.05, the differential expressed mRNAs (DEMs) in LSCC were screened with GEO2R (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html), an interactive software for identifying the ectopic genes in different groups [15]. Followed, the significant DEMs (adjust P<0.05 and log2 (fold change) ≤1 or ≥-1)) were further screened manually.
Creating of protein-protein interaction networks and identifying of hub-genes: After acquiring the DEMs of each included profile, the intersections of all included profiles were calculated by an online software, Venn Diagrams (http://bioinformatics.psb.ugent.be/beg/tools/venn-diagrams). Based on the intersections, the protein-protein interaction (PPI) networks for up- and down-regulated DEMs were created with Cytoscape (version 3.6.0) [16] and STRING (version 11.0) (https://string-db.org) [17]. Followed the construction of PPI networks, hub-genes were screened through a Cytoscape plugin cytoHubba [18]. Hub-genes were defined as the top 10 of node scores.
Acquiring of the mRNA profile for verification of hub-genes: For verifying the expression levels of hub-genes, a novel mRNA profile was achieved from The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) [19], through SangerBox (version 1.0.9) (http://soft.sangerbox.com/), an integrated bioinformatics analysis software, which can be used for data acquisition from TCGA conveniently. Procedure: launch TCGA downloading tool (version 16) by clicking on the icon, double click “Head and Neck Squamous Cell Carcinoma (TCGA-HNSC)” on the left of dialog, select “all transcriptome data” in the pop-up dialog, select “HTSeq-FPKM-UQ (544)” in the drop-down. Use the same procedure to download clinical follow-up data. After acquiring the mRNA profile from TCGA, the expression values of hub-genes were calculated as log2 (TPM+1), (TPM, transcripts per million) [20].
Analyzing of functional enrichment for DEMs: For exploring the biological functions and possible mechanisms of DEMs, especially for hub-genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were completed with The Database for Annotation, Visualization and Integrated Discovery (DAVID) database (version 6.8) (https://david.ncifcrf.gov), an interactive project [21].
Evaluation of the clinical implications for hub-genes: Receiver operating characteristic (ROC) curve and area under ROC curve (AUC) were employed for evaluating the diagnosis and prognosis potential, respectively. The Drug and Gene Interaction Database (DGIdb) (https://dgidb.genome.wustl.edu/) were used to identify the drugs interacted with some hub-genes [22].
Statistical analysis
Independent sample t-test (normal distribution data) or Mann-Whitney U test (non-normal distribution data) was used for measuring the expression levels of hub-genes. Chi square test was used for overall survival analysis. SPSS 22.0 software (IBM SPSS, USA) and GraphPad Prism 7 software (GraphPad Software Inc., USA) were used for statistical analysis and the vi sualization of results. P<0.05> indicates a significant difference.
Results
LSCC related mRNA expression profiles in GEO DataSets:According to the retrieval filters and search terms, total seven expression profiles were preliminarily retrieved (Retrieval date May 04, 2020). Of them, GSE88862 (a de facto lung squamous cell carcinoma profile, also abbreviated as LSCC), and GSE148944 (an oral tongue squamous cell carcinoma profile) were firstly removed by browsing the titles. After that, GSE59102 and GSE123986, deriving from unpaired LSCC tissues, were further excluded by learning the research methods. Ultimately, GSE51985, GSE84957, and GSE143224, meeting the screening criteria, were included in the subsequent analyses (Figure 1). Of the above three profiles, GSE51985 with 10 paired LSCC samples, was submitted on November 02, 2013 and updated on August 22, 2019 [23]. GSE84957 with nine paired samples, was submitted on July 28, 2016 and updated on March 30, 2017 [24]. And, GSE143224 with 14 paired samples, was submitted on January 07, 2020 and updated on March 23, 2020 [25].
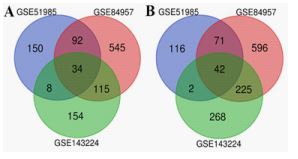
Distribution of DEMs was not exactly identical in different expression profiles: After screening the qualified expression profiles, the DEMs for each profile was preliminarily screened and downloaded with GEO2R according to the default parameters. Secondly, the DEMs were further selected by manually following the threshold (adjust P<0.05> and log2 (fold change) ≥1 or ≤-1). Finally, total 515, 1720, and 854 DEMs were screened out from GSE51985, GSE84957, and GSE143224, respectively. Added, the intersections of the above three profiles were calculated with Venn diagram. Accordingly, total 34 up-regulated and 42 down-regulated DEMs were included in the intersections (Figure 1, 2A~B).e
PPI networks showed that the interaction among the hub-genes was complicated: Considering the intersection of the up- or the down-regulated DEMs, the PPI networks were construct¬ed with Cytoscape separately. The network diagrams showed that the interactions of the DEMs were complicated, especially for up-regulated DEMs (Figure S1). Further, the top 10 of the node scores were extracted with cytoHubba and defined as the hub-genes. The up-regulated hub-genes were as follows: LAMC2, FOXM1, COL4A2, SNAI2, MMP3, SPP1, PLAUR, IGFBP3, MMP1, and PLAU. The down-regulated hub-genes were as fol¬lows: FUT6, EPHX2, ATP6V0A4, CYP2J2, ST6GALNAC1, ST3GAL4, GALNT12, MUC1, GCNT3, and LEPR (Figure 3A~B).
Evaluating of the expression values for the hub-genes in TCGA: For verifying the expression tendency of the hub-genes, a novel mRNA expression profile associated with head and neck squamous cell carcinoma was downloaded from TCGA by SangerBox, consisting of 500 cancerous samples and 44 normal samples (Retrieval date May 06, 2020). For improving the accuracy and pertinence of the analyses, the data related to LSCC was further selected manually, consisting of 111 cancerous samples and 12 normal samples. All of the hub-genes expression levels in TCGA were consistent with that in GEO profiles (Figure 4A~B).
Hub-genes participated in plenty of biological processes and pathways of LSCC: For analyzing the biological functions of DEMs, especially for hub-genes, GO analysis showed that hub-genes were involved in various biological processes (extracellular matrix disassembly/organization, O-glycan processing, or protein glycosylation) and multiple molecular functions (serine-type endopeptidase activity, metalloendopeptidase activity, or toxic substance binding) as diverse cellular components (extracellular region/matrix/space, extracellular exosome, or apical plasma membrane) (Retrieval date May 06, 2020) (Table 1~2). KEGG pathway analyses presented that hub-genes participated in multiple pathways (PI3K-Akt pathway, Pathways in cancer, or Metabolic pathways) (Retrieval date May 06, 2020) (Table 1~2).
Diagnosis and prognosis values of hub-genes associated with PI3K-Akt signaling pathway: Prior researches have demonstrated that PI3K-Akt pathway effects on the initiation and progression of LSCC [13,26-28]. Based on the previous findings, the present study further analyzed the potential clinical values of COL4A2, LAMC2, and SPP1, which enriched in PI3K-Akt signaling pathway. ROC curve analyses demonstrated that COL4A2, LAMC2, and SPP1 presented potential diagnostic values with 0.993, 0.911, and 0.951 AUC (P<0.001), respectively (Figure 5A). Unfortunately, the overall survival analyses showed no significant correlations between the expression levels of COL4A2, LAMC2, and SPP1 and the overall survival times (P>0.05) (Figure 5B). Nevertheless, patients showing high expression values of LAMC2 or SPP1 seemingly survived shorter than patients showing low values based on the present data.
Hub-genes participated in plenty of biological processes and pathways of LSCC: For analyzing the biological functions of DEMs, especially for hub-genes, GO analysis showed that hub-genes were involved in various biological processes (extracellular matrix disassembly/organization, O-glycan processing, or protein glycosylation) and multiple molecular functions (serine-type endopeptidase activity, metalloendopeptidase activity, or toxic substance binding) as diverse cellular components (extracellular region/matrix/space, extracellular exosome, or apical plasma membrane) (Retrieval date May 06, 2020) (Table 1~2). KEGG pathway analyses presented that hub-genes participated in multiple pathways (PI3K-Akt pathway, Pathways in cancer, or Metabolic pathways) (Retrieval date May 06, 2020) (Table 1~2).
Diagnosis and prognosis values of hub-genes associated with PI3K-Akt signaling pathway: Prior researches have dem¬onstrated that PI3K-Akt pathway effects on the initiation and progression of LSCC [13,26-28]. Based on the previous find¬ings, the present study further analyzed the potential clinical values of COL4A2, LAMC2, and SPP1, which enriched in PI3K-Akt signaling pathway. ROC curve analyses demonstrated that COL4A2, LAMC2, and SPP1 presented potential diagnostic val¬ues with 0.993, 0.911, and 0.951 AUC (P<0.001), respectively (Figure 5A). Unfortunately, the overall survival analyses showed no significant correlations between the expression levels of CO¬L4A2, LAMC2, and SPP1 and the overall survival times (P>0.05) (Figure 5B). Nevertheless, patients showing high expression val¬ues of LAMC2 or SPP1 seemingly survived shorter than patients showing low values based on the present data.
Therapeutic medications interacted with COL4A2, LAMC2, and SPP1: For identifying the therapeutic values of COL4A2, LAMC2, and SPP1, the three hub-genes were submitted into DGIdb. The results presented that some drugs may interacted with the above-mentioned three hub-genes (Retrieval data Dec. 10, 2020) (Table 3).
Discussion
As a common malignant tumor of head and neck, the researches on LSCC have obtained remarkable achievements, covering laboratory and clinical researches [6,7,29,30]. However, the pathogenesis of LSCC has not been well elucidated. The specific tumor markers and effective molecular targets are still deficient. Considering the above-mentioned, the present study screened out the hub-genes that exerted pivotal molecular functions in the initiation and progression of LSCC through bioinformatics analyses. The detailed analyses demonstrated that plentiful aberrantly expressed mRNAs consisted in LSCC. The DEMs, especially the hub-genes, independently or synergistically participated in the occurrence and development of LSCC, through diversity molecular functions and signaling pathways. For instance, the functional enrichment analyses and PPI networks showed that the up-regulated hub-genes, COL4A2 and LAMC2, were synergistically involved in the regulation of PI3K-Akt pathway, Pathways in cancer, and ECM-receptor interaction. Even, COL4A2 and LAMC2 presented significant diagnostic values and potential prognostic values.
Expression profile analyses were extensively applied to the researches on diversity cancers, including nasopharyngeal carcinoma [31], hepatocellular carcinoma [32], osteosarcoma [33], cholangiocarcinoma [34], and LSCC [35,36]. In addition to the original researches, the re-analyses of expression profiles also presented significant roles in tumor related studies [37-39]. For instance, TCGA database was adopted to discern the relationship between cellular heterogeneity and the prognosis of LSCC patients [40]. Added, TCGA combined with bioinformatics analyses proposed the significant prognostic value of long non-coding RNAs for LSCC [41] and speculated the possibility of the homeobox A cluster gene family as specific targets of LSCC targeted therapy [42]. Furthermore, a weighted gene co-expression analysis of GEO profile distinguished the hub-genes participating in the evolution of LSCC [43].
In view of previous research methods and achievements, the present study proceeded from screening the DEMs of LSCC through mining expression profiles of LSCC in GEO Data Sets. For improving the reliability and veracity of present analysis, the current study extracted the DEMs from three independent profiles and brought the intersections of the three groups of DEMs into subsequent analyses. The DEMs, simultaneously al-tered in certain cancerous tissues, may exert synergistic or an¬tagonistic effects on the cellular biological processes [44, 45]. Consistently, PPI networks and functional enrichment analyses of the present study demonstrated that multiple up-regulated or down-regulated DEMs, especially the hub-genes, enriched in the identical pathway or GO term. For instance, 14 up-regulated DEMs, including seven hub-genes, concurrently enriched in the common GO term, extracellular region (GO: 0005576). Mean¬ while, the analysis also indicated that 10 down-regulated DEMs, including eight hub-genes, simultaneously enriched in the met¬abolic pathways (has 01100). The above-mentioned phenom¬ena suggested that the hub-genes, concurrently located in the same GO term or pathway, exerted similar or opposite effects on the identical biological behavior.
Table 1: Functional enrichment analysis of up-regulated miRNAs.
| Category | Count | P-value | Genes |
|---|---|---|---|
| GOTERM-CC | |||
| GO:0005576~extracellular region | 14 | 1.33×10-6 | COL4A2, COL4A1, STC2, FST, MMP3, MMP1, MMP11, PTHLH, LAMB3, TGFBI, LAMC2, IGFBP3, PLAU, SPP1 |
| GO:0031012~extracellular matrix | 5 | 0.001714 | COL4A2, COL4A1, TGFBI, MMP1, MMP11 |
| GO:0005615~extracellular space | 9 | 0.001848 | PTHLH, STC2, TGFBI, LAMC2, MMP3, IGFBP3, SERPINH1, PLAU, SPP1 |
| GO:0048471~perinuclear region of cytoplasm | 6 | 0.004246 | LAMP3, STC2, MYO1B, CLIC4, LAMC2, SPP1 |
| GO:0005788~endoplasmic reticulum lumen | 4 | 0.004561 | COL4A2, COL4A1, SERPINH1, PLAUR |
| GO:0005587~collagen type IV trimer | 2 | 0.010491 | COL4A2, COL4A1 |
| GO:0005578~proteinaceous extracellular matrix | 4 | 0.011387 | TGFBI, MMP3, MMP1, MMP11 |
| GO:0070062~extracellular exosome | 11 | 0.018709 | WARS, COL4A2, KRT17, MYO1B, CLIC4, TGFBI, IGFBP3, SERPINH1, PLAU, SPP1, PLAUR |
| GOTERM-MF | |||
| GO:0004252~serine-type endopeptidase activity | 4 | 0.010235 | MMP3, MMP1, PLAU, MMP11 |
| GO:0004222~metalloendopeptidase activity | 3 | 0.017098 | MMP3, MMP1, MMP11 |
| GO:0050840~extracellular matrix binding | 2 | 0.045227 | TGFBI, SPP1 |
| GOTERM-BP | |||
| GO:0022617~extracellular matrix disassembly | 6 | 2.17×10-7 | LAMB3, LAMC2, MMP3, MMP1, SPP1, MMP11 |
| GO:0030574~collagen catabolic process | 5 | 4.88×10-6 | COL4A2, COL4A1, MMP3, MMP1, MMP11 |
| GO:0030198~extracellular matrix organization | 6 | 2.31×10-5 | COL4A2, LAMB3, COL4A1, TGFBI, LAMC2, SPP1 |
| GO:0008544~epidermis development | 4 | 4.60×10-4 | PTHLH, LAMB3, KRT17, LAMC2 |
| GO:0001525~angiogenesis | 4 | 0.007203 | WARS, COL4A2, CLIC4, TGFBI |
| GO:0051798~positive regulation of hair follicle development | 2 | 0.010673 | KRT17, FST |
| GO:0038063~collagen-activated tyrosine kinase receptor signaling pathway | 2 | 0.010673 | COL4A2, COL4A1 |
| GO:0071711~basement membrane organization | 2 | 0.014206 | COL4A1, MMP11 |
| GO:0001649~osteoblast differentiation | 3 | 0.014762 | SNAI2, IGFBP3, SPP1 |
| GO:0044267~cellular protein metabolic process | 3 | 0.018735 | TGFBI, IGFBP3, MMP1 |
| KEGG PATHWAY | |||
| hsa04512:ECM-receptor interaction | 5 | 3.87×10-5 | COL4A2, LAMB3, COL4A1, LAMC2, SPP1 |
| hsa05222:Small cell lung cancer | 4 | 9.08×10-4 | COL4A2, LAMB3, COL4A1, LAMC2 |
| hsa04510:Focal adhesion | 5 | 0.001071 | COL4A2, LAMB3, COL4A1, LAMC2, SPP1 |
| hsa05146:Amoebiasis | 4 | 0.001721 | COL4A2, LAMB3, COL4A1, LAMC2 |
| hsa04151:PI3K-Akt signaling pathway | 5 | 0.006998 | COL4A2, LAMB3, COL4A1, LAMC2, SPP1 |
| hsa05200:Pathways in cancer | 5 | 0.011028 | COL4A2, LAMB3, COL4A1, LAMC2, MMP1 |
Table 2: Functional enrichment analysis of down-regulated miRNAs.
| Category | Count | P-value | Genes |
|---|---|---|---|
| GOTERM-CC | |||
| GO:0070062~extracellular exosome | 20 | 1.07×10-6 | EVL |
| 3 | 51 | melena | MUC1, C7, GCNT3, CAPN5, CAB39L, CYP2J2, FUT6, FAM3B, IL1RN, EPHX2, UBL3, GPD1L, GGT6, COL14A1, ST3GAL4, NUCB2, SH3BGRL2, NDRG2, ATP6V0A4, CFD |
| GO:0016324~apical plasma membrane | 4 | 0.024118 | MUC1, SORBS2, STK39, ATP6V0A4 |
| GO:0005794~Golgi apparatus | 6 | 0.035755 | ST3GAL4, FUT6, HSPB8, NUCB2, NDRG2, ST6GALNAC1 |
| GO:0000139~Golgi membrane | 5 | 0.036767 | GCNT3, ST3GAL4, FUT6, GALNT12, ST6GALNAC1 |
| GOTERM-MF | |||
| GO:0015643~toxic substance binding | 2 | 0.023911a | EPHX2, CYP4B1 |
| GO:0008373~sialyltransferase activity | 2 | 0.037602 | ST3GAL4, ST6GALNAC1 |
| GO:0005125~cytokine activity | 3 | 0.048769 | FAM3D, FAM3B, IL1RN |
| GOTERM-BP | |||
| GO:0016266~O-glycan processing | 4 | 2.62×10-4 | MUC1, GCNT3, ST3GAL4, GALNT12 |
| GO:0006486~protein glycosylation | 3 | 0.023113 | ST3GAL4, FUT6, ST6GALNAC1 |
| GO:0006957~complement activation, alternative pathway | 2 | 0.026769 | C7, CFD |
| GO:0019373~epoxygenase P450 pathway | 2 | 0.036879 | CYP2J2, EPHX2 |
| GO:0001574~ganglioside biosynthetic process | 2 | 0.036879 | ST3GAL4, ST6GALNAC1 |
| GO:0097503~sialylation | 2 | 0.040894 | ST3GAL4, ST6GALNAC1 |
| KEGG PATHWAY | |||
| hsa00512:Mucin type O-Glycan biosynthesis | 3 | 0.003550 | GCNT3, GALNT12, ST6GALNAC1 |
| hsa01100:Metabolic pathways | 10 | 0.004317 | GCNT3, GGT6, CYP2J2, ST3GAL4, FUT6, EPHX2, ATP6V0A4, GALNT12, ST6GALNAC1, ACOX3 |
GO: gene ontology, CC: cellular components, MF: molecular functions, BP: biological processes, KEGG: Kyoto Encyclopedia of Genes and Genomes. The red indicates the hub-genes.
Table 3: Drugs interacted with COL4A2, LAMC2, and SPP1.
| Gene | Drug | Interaction types | Sources | Interaction score | PubMed ID |
|---|---|---|---|---|---|
| COL4A2 | Collagenase Clostridium Histolyticum | N/A | ChemblInteractions | 0.68 | None found |
| COL4A2 | Navoximod | N/A | TTD | 6.31 | None found |
| COL4A2 | Ocriplasmin | N/A | ChemblInteractions | 0.49 | None found |
| LAMC2 | Ocriplasmin | N/A | ChemblInteractions | 1.46 | None found |
| SPP1 | ASK-8007 | Inhibitor | ChemblInteractions | 9.47 | None found |
| SPP1 | Calcitonin | N/A | NCI | 3.16 | 8013390 |
| SPP1 | Alteplase | N/A | NCI | 0.9 | 12009309 |
| 8 | 73 | melena | EVL | ||
| SPP1 | Wortmannin | N/A | NCI | 0.51 | 14703434 |
| SPP1 | Gentamicin | N/A | NCI | 0.82 | 11274264 |
| SPP1 | Tacrolimus | N/A | NCI | 0.56 | 16103732 |
ChemblInteractions: The ChEMBL Bioactivity Database, TTD: Therapeutic Target Database, NCI: NCI Cancer Gene Index.
Remarkably, of the enriched pathways, PI3K-Akt pathway, a pivotal pathway regulating epithelial-mesenchymal transition [13,46], covered three hub-genes (COL4A2, LAMC2, SPP1). Consequently, the present study speculated that the three hub-genes may affect the initiation and progression of LSCC. The further analyses suggested that all of the three hub-genes presented significant diagnostic values with more than 0.9 AUC. Moreover, the prognostic analyses suggested that the patients showing low expression values of LAMC2 or SPP1 apparently presented the better prognosis, though the statistic difference was not significant. In addition, the present analyses also explored the therapeutic agents, such as Navoximod, Ocriplasmin, and ASK-8007, which may interact with COL4A2, LAMC2 or SPP1. The above finding furnished a valuable reference for subsequent research.
Herein, the present study screened 34 up- and 42 down-regulated DEMs through mining expression profiles. Of the DEMs, 10 up- and 10 down-regulated hub-genes were defined through a series of bioinformatics analyses. Furthermore, detailed analyses suggested that the hub-genes playing pivotal roles in the initiation and progression of LSCC through PI3K-Akt pathway, pathways in cancer, and metabolic pathways. Even, as the members of PI3K-Akt pathway, the hub-genes, LAMC2 and SPP1, presented the significant diagnostic values and potential prognostic values. In addition, some drugs, ASK-8007, Ocriplasmin or Calcitonin may interact with LAMC2 or SPP1. The present study furnished significant references for the further researches on exploring the pathogenesis and targeted therapy of LSCC.
Declarations
Author contributions: Conceptualization, Data curation, Methodology, and Validation, YUAN LIANG and LEI ZHAO; Writing-original draft, YUAN LIANG; Writing - review & editing and Project administration, LEI ZHAO.
Ethical compliance: Not applicable.
Conflicts of Interest: There are no conflicts to declare.
Acknowledgements: All of the analyses benefited from the free public databases, as follows: GEO, TCGA, Cytoscape, STRING, DIVID and DGIdb. This study was funded by Hebei Medical Science Research Program [NO. 20200575]; Medical Science Foundation of Hebei University [NO. 2021B16]; Foundation Project of Affiliated Hospital of Hebei University [NO. 2019Q029].
References
- Ciolofan MS, Vlăescu AN, Mogoantă CA, Ioniță E, Ioniță I, Căpitănescu AN, et al. Clinical, Histological and Immunohistochemical Evaluation of Larynx Cancer. Curr Health Sci J. 2017; 43(4): 367-375.
- Nocini R, Molteni G, Mattiuzzi C, Lippi G. Updates on larynx cancer epidemiology. Chin J Cancer Res. 2020; 32(1): 18-25.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66(1): 7-30.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020; 70(1): 7-30.
- Vaiciulis P, Liutkeviciene R, Liutkevicius V, Vilkeviciute A, Gedvilaite G, Uloza V. Association of Relative Leucocyte Telomere Length and Gene Single Nucleotide Polymorphisms (TERT, TRF1, TNKS2) in Laryngeal Squamous Cell Carcinoma. Cancer Genomics Proteomics. 2020; 17(4): 431-439.
- Li X, Xu F, Meng Q, Gong N, Teng Z, Xu R, et al. Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis. Cell Death Dis. 2020; 11(6): 472.
- Alessandrini L, Franz L, Ottaviano G, Ghi MG, Lanza C, Blandamura S, et al. Prognostic role of programmed death ligand 1 (PD-L1) and the immune microenvironment in laryngeal carcinoma. Oral Oncol. 2020; 108: 104836.
- Gao W, Zhang C, Li W, Li H, Sang J, Zhao Q, et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Mol Ther. 2019; 27(2): 365-379.
- Wu Y, Zhang Y, Zheng X, Dai F, Lu Y, Dai L, et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol Cancer. 2020; 19(1): 99.
- Wang X, Yu B, Jin Q, Zhang J, Yan B, Yang L, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA RP11-159K7.2/miR-206/DNMT3A axis. J Cell Mol Med. 2020; 24(12): 6781-6795.
- Zhao R, Tian L, Zhao B, Sun Y, Cao J, Chen K, et al. FADS1 promotes the progression of laryngeal squamous cell carcinoma through activating AKT/mTOR signaling. Cell Death Dis. 2020; 11(4): 272.
- Liu L, Zhang P, Shao Y, Quan F, Li H. Knockdown of FOXJ1 inhibits the proliferation, migration, invasion, and glycolysis in laryngeal squamous cell carcinoma cells. J Cell Biochem. 2019; 120(9): 15874-15882.
- Zhu Y, Yan L, Zhu W, Song X, Yang G, Wang S. MMP2/3 promote the growth and migration of laryngeal squamous cell carcinoma via PI3K/Akt-NF-κB-mediated epithelial-mesenchymal transformation. J Cell Physiol. 2019.
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41(Database issue): D991-5, 2013.
- Davis, S. and P.S. Meltzer, GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics, 2007; 23(14): p. 1846-7.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13(11): 2498-504.
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47(D1): D607-D613.
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. (Suppl 4):S11, 2014.
- Gao GF, Parker JS, Reynolds SM, Silva TC, Wang LB, Zhou W, et al. Before and After: Comparison of Legacy and Harmonized TCGA Genomic Data Commons’ Data. Cell Syst. 2019; 9(1): 24-34.
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45(W1): W98-W102.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 2009; 4(1): p. 44-57.
- Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018; 46(D1): D1068-D1073.
- Lian M, Fang J, Han D, Ma H, Feng L, Wang R, et al. Microarray gene expression analysis of tumorigenesis and regional lymph node metastasis in laryngeal squamous cell carcinoma. PLoS One. 2013; 8(12): e84854.
- Feng L, Wang R, Lian M, Ma H, He N, Liu H, et al. Integrated Analysis of Long Noncoding RNA and mRNA Expression Profile in Advanced Laryngeal Squamous Cell Carcinoma. PLoS One. 2016; 11(12): e0169232.
- Nicolau-Neto P, de Souza-Santos PT, Severo Ramundo M, Valverde P, Martins I, Santos IC, et al. Transcriptome Analysis Identifies ALCAM Overexpression as a Prognosis Biomarker in Laryngeal Squamous Cell Carcinoma. Cancers (Basel). 2020; 12(2): 470.
- Tang T, Xiao ZY, Shan G, Lei HB. Descending-SHIP2-mediated radiosensitivity enhancement through PI3K/Akt signaling pathway in laryngeal squamous cell carcinoma. Biomed Pharmacother. 2019; 118: 109392.
- Ni HS, Hu SQ, Chen X, Liu YF, Ni TT, Cheng L. Tra2β silencing suppresses cell proliferation in laryngeal squamous cell carcinoma via inhibiting PI3K/AKT signaling. Laryngoscope. 2019; 129(9): E318-E328.
- Ye D, Zhou C, Deng H, Lin L, Zhou S. MicroRNA-145 inhibits growth of laryngeal squamous cell carcinoma by targeting the PI3K/Akt signaling pathway. Cancer Manag Res. 2019; 11: 3801-3812.
- Shi Y, Zhou L, Tao L, Zhang M, Chen XL, Li C, et al. Management of the N0 neck in patients with laryngeal squamous cell carcinoma. Acta Otolaryngol. 2019; 139(10): 908-912.
- Cayonu M, Tuna EU, Acar A, Dinc ASK, Sahin MM, Boynuegri S, et al. Lymph node yield and lymph node density for elective level II-IV neck dissections in laryngeal squamous cell carcinoma patients. Eur Arch Otorhinolaryngol. 2019; 276(10): 2923-2927.
- Chen S, Youhong T, Tan Y, He Y, Ban Y, Cai J, et al. EGFR-PKM2 signaling promotes the metastatic potential of nasopharyngeal carcinoma through induction of FOSL1 and ANTXR2. Carcinogenesis. 41(6):723-733, 2020.
- Midorikawa Y, Yamamoto S, Tatsuno K, Renard-Guillet C, Tsuji S, Hayashi A, et al. Accumulation of Molecular Aberrations Distinctive to Hepatocellular Carcinoma Progression. Cancer Res. 2020; 80(18): 3810-3819.
- Raimondi L, De Luca A, Gallo A, Costa V, Russelli G, Cuscino N, et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis. 2020; 41(5): 666-677.
- Zhou Q, Lin M, Feng X, Ma F, Zhu Y, Liu X, et al. Targeting CLK3 inhibits the progression of cholangiocarcinoma by reprogramming nucleotide metabolism. J Exp Med. 2020; 217(8): e20191779.
- Lyu K, Li Y, Xu Y, Yue H, Wen Y, Liu T, et al. Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma. RNA Biol. 2020; 17(7): 977-989.
- Zhang, F. and H. Cao, MicroRNA‑143‑3p suppresses cell growth and invasion in laryngeal squamous cell carcinoma via targeting the k‑Ras/Raf/MEK/ERK signaling pathway. International journal of oncology, 2019; 54(2): p. 689-701.
- Sun T, Bi F, Liu Z, Yang Q. SLC7A2 serves as a potential biomarker and therapeutic target for ovarian cancer. Aging (Albany NY). 2020; 12(13): 13281-13296.
- Ruckert MT, Brouwers-Vos AZ, Nagano LFP, Schuringa JJ, Silveira VS. HUWE1 cooperates with RAS activation to control leukemia cell proliferation and human hematopoietic stem cells differentiation fate. Cancer Gene Ther. 2020; 27(10-11): 830-833.
- Shin WS, Park MK, Lee YH, Kim KW, Lee H, Lee ST. The catalytically defective receptor protein tyrosine kinase EphA10 promotes tumorigenesis in pancreatic cancer cells. Cancer Sci. 2020; 111(9): 3292-3302.
- Song L, Zhang S, Yu S, Ma F, Wang B, Zhang C, et al. Cellular heterogeneity landscape in laryngeal squamous cell carcinoma. Int J Cancer. 147(10):2879-2890, 2020.
- Gong S, Xu M, Zhang Y, Shan Y, Zhang H. The Prognostic Signature and Potential Target Genes of Six Long Non-coding RNA in Laryngeal Squamous Cell Carcinoma. Front Genet. 2020; 11:413.
- Li, J., M. Ye, and C. Zhou, Expression Profile and Prognostic Values of Family Members in Laryngeal Squamous Cell Cancer. Frontiers in oncology, 10: p. 368, 2020.
- Zhang H, Zhao X, Wang M, Ji W. Key modules and hub genes identified by coexpression network analysis for revealing novel biomarkers for larynx squamous cell carcinoma. J Cell Biochem. 2019; 120(12): 19832-19840.
- Komura N, Mabuchi S, Shimura K, Yokoi E, Kozasa K, Kuroda H, et al. The role of myeloid-derived suppressor cells in increasing cancer stem-like cells and promoting PD-L1 expression in epithelial ovarian cancer. Cancer Immunol Immunother. 2020; 69(12): 2477-2499.
- He J, Wu M, Xiong L, Gong Y, Yu R, Peng W, et al. BTB/POZ zinc finger protein ZBTB16 inhibits breast cancer proliferation and metastasis through upregulating ZBTB28 and antagonizing BCL6/ZBTB27. Clin Epigenetics. 2020; 12(1): 82.
- Yuan Y, Liao H, Pu Q, Ke X, Hu X, Ma Y, et al. miR-410 induces both epithelial-mesenchymal transition and radioresistance through activation of the PI3K/mTOR pathway in non-small cell lung cancer. Signal Transduct Target Ther. 2020; 5(1): 85.